snpEff tutorial | Installation | variant calling
Автор: Study Tech
Загружено: 2025-12-28
Просмотров: 71
SnpEff
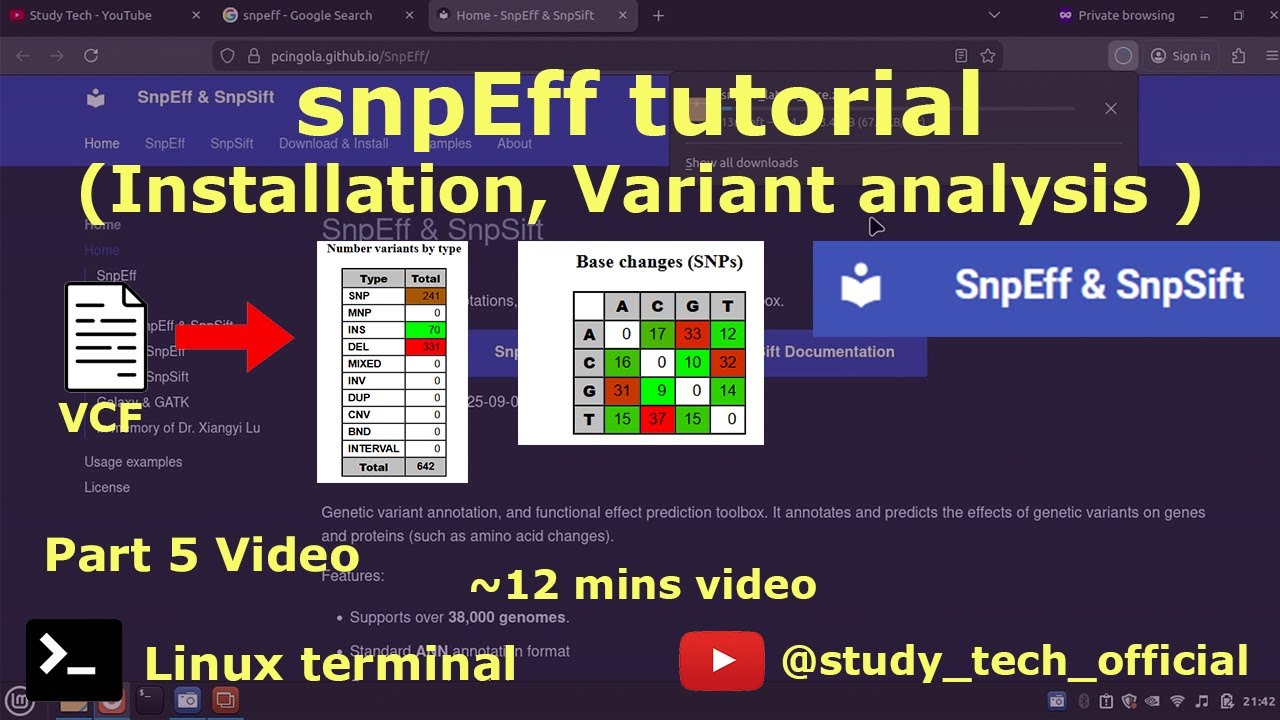
Genetic variant annotation, and functional effect prediction toolbox. It annotates and predicts the effects of genetic variants on genes and proteins (such as amino acid changes).
1️⃣ Search "snpEff"
https://pcingola.github.io/SnpEff/
2️⃣ Download the file
snpEff_latest_core.zip
3️⃣ Unzip and Check
java -jar snpEff.jar
4️⃣ Database information
java -jar snpEff.jar databases database.txt
5️⃣ Now search for "Saccharomyces cerevisiae"
Saccharomyces_cerevisiae
6️⃣ Download the data
java -jar snpEff.jar download Saccharomyces_cerevisiae
6️⃣ Run
java -Xmx8g -jar snpEff.jar -v -canon Saccharomyces_cerevisiae -csvStats result AS_W_ETOH_calls.vcf AS_W_ETOH_calls_annotate.vcf
#### An additional parameter to add to our command is Xmx8G, a Java parameter to define available memory.
✉️Email- graviti.work@outlook.com
Email me for further contact
📢 Save my video URL on a notepad in your lab computer. So that you can get this video
📢 Subscribe to my Study tech youtube channel. Because it is free for you but it will help me a lot.
====================== Follow genomic variants analysis pipeline =====================
------ Tutorial made in the year 2024-2025
🎯Step 1: Download fastq(raw file-SRA) file from NCBI
• How to download fastq file | Full tutorial...
🎯Step 2: Fastq file quality check [Fastqc tool]
• How to install fastqc in windows | Beginne...
🎯Step 3: Trim the adapter and bad reads from the fastq file [Fastp tool]
• Fastp | Install | Run | Paired end #bioinf...
🎯Step 4: Mapping/Alignment of fastq to generate SAM file [STAR tool]
• STAR | Install | Build index | #bioinforma...
• STAR | sequence alignment | mapping #bioi...
🎯Step 5: Sort the SAM file into BAM [ Samtools ]
• Samtools | Install | SAM to BAM conversion...
🎯Step 6: AddOrReplaceReadGroup of BAM files [GATK tools]
• GATK Installation | AddOrReplaceReadGroup...
🎯Step 7: Mark Duplication of BAM files [GATK tools]
• GATK Tutorial | Mark Duplication | Varian...
🎯Step 8: Merge BAM files [ Samtools ]
• BCFtools installation | Merge BAM files #b...
🎯Step 9: mpileup for variant calling [ BCFtools ]
• BCFtools tutorial | mpileup | variant call...
🎯Step 10: snp analysis [snpEFF tools]
• snpEff tutorial | Installation | variant c...
====================== Subscribe to my channel, and learn complete genomics analysis 💻 System configuration
Procession- Intel i5 4th gen, 1.7 GHz, 4 CPU(or thread)
RAM-8GB
Boot and working drive- 256GB SSD
OS- Linux Mint 21.1 Cinnamon
📢 Guys, if you need any tutorial or help regarding Bioinformatics work, we will try to help you by Video or Chat.
Fill free to ask in the comment section.
If you are very new to Bioinformatics and have some very basic and silly questions. You are most welcome. Don't hesitate to ask us.
You can write in the comment section.
=========== Jump to
00:05 Introduction
00:54 Installation of snpEff
03:20 View snpEff database
05:37 Download snpEff database
07:18 Annotation vcf in to result
10:16 Description of snpEff result
🏷️ Video specific tags
snpeff,
snpeff install,
snpeff tutorial,
snpeff database,
snpeff database download,
snp analysis,
indel analysis,
snp,
variant calling,
variant analysis,
how to install snpeff,
genomics analysis,
haplotype,
deletion, duplication,
variant calls,
vcf, bcf, variant call format,
snp analysis, snp call, deletion analysis,
🏷️ Channel tags
transcriptomics, genomics, RNAseq, DNAseq,fasta, fastq, bam, sam,
quality control and trimming,
bioinformatic tutorial, tutorial,bioinfo,
tool, software, package, linux, ubuntu
synopsis, script, code, terminal,prompt, command,
sequence alignment, sequence, bioinformatics training, file format, bioinformatics projects,
#ncbi #genomics #beginners #tutorial #howto #omics #research #biology #ngs #python
Доступные форматы для скачивания:
Скачать видео mp4
-
Информация по загрузке:

















![Как внимание стало настолько эффективным [GQA/MLA/DSA]](https://image.4k-video.ru/id-video/Y-o545eYjXM)

