Foundation Models in Genomics: A Supervised Alternative to DNA Language Model: Asa Ben-Hur, 28/07/25
Автор: TIA Warwick
Загружено: 2025-07-28
Просмотров: 282
TIA Centre Seminar Series: Prof Asa Ben-Hur
Full Title: Foundation Models in Genomics: A Supervised Alternative to DNA Language Models
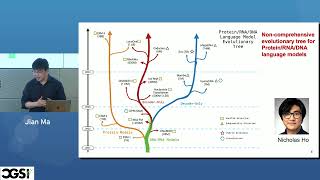
Abstract: DNA language models—self-supervised models trained purely on nucleotide sequences—have emerged in recent years as a platform for building deep learning models in genomics. However, there is a compelling alternative: supervised chromatin state models trained on large-scale epigenomic data to predict regulatory activity directly from sequence. These models can be trained efficiently on modest hardware, making them more accessible and easier to deploy. More importantly, they offer superior predictive accuracy and greater interpretability compared to DNA language models. In this talk, I will compare these two paradigms and argue that supervised chromatin state models are a more practical and powerful option for many regulatory genomics applications. I will illustrate this by applying both approaches to the prediction of active enhancers, gene expression, and alternative splicing in both plant and human systems.

Доступные форматы для скачивания:
Скачать видео mp4
-
Информация по загрузке: